Biophysical Data Analysis and Reasoning with Edison Analysis Agent
Asmamaw (Oz) Wassie, Michaela Hinks
Date:
02.13.2026
Technologies that probe the biophysical properties of biomolecules are foundational to modern biology, enabling direct measurement of structure, dynamics, and molecular interactions. Single-molecule sensing with nanopores enables direct, label-free measurement of individual biomolecules in real time, rather than averaging signals across millions of molecules as most bulk assays do. Such a capability would be transformative as it allows for precision medicine and diagnostics, scalable drug discovery, rapid pathogen surveillance, protein characterization, and sensitive analysis of chemicals. Recent advances in nanopore devices have expanded the capabilities of nanopores to the single-molecule level, opening the door to quantitative characterization of small biomolecules such as drug compounds, DNA, RNA, and peptides. In a recent pioneering work, Zhang et al. developed a custom-engineered nanopore platform capable of trapping single amino acid molecules and sensitively resolving their characteristic ionic current blockade profiles. We reasoned that the rich time-series data generated by this system would provide a stringent test case for the Edison Analysis Agent’s ability to autonomously explore timeseries biophysical signals. Here, we demonstrate that the Edison Analysis agent can recapitulate a central finding of Zhang et al., that amino acid molecular volume is the primary determinant of ionic current blockade amplitude within this nanopore configuration, through systematic analysis and reasoning over this nanopore recording dataset.
Research Objective and Nanopore Dataset
To evaluate whether Edison Analysis could interpret nanopore time-series data and to initiate the exploration of this dataset, we provided the agent with a brief prompt (Figure 1A) and a csv file of nanopore recording values for individual amino acids (Figure 1B) pulled from the supplementary materials of the manuscript.
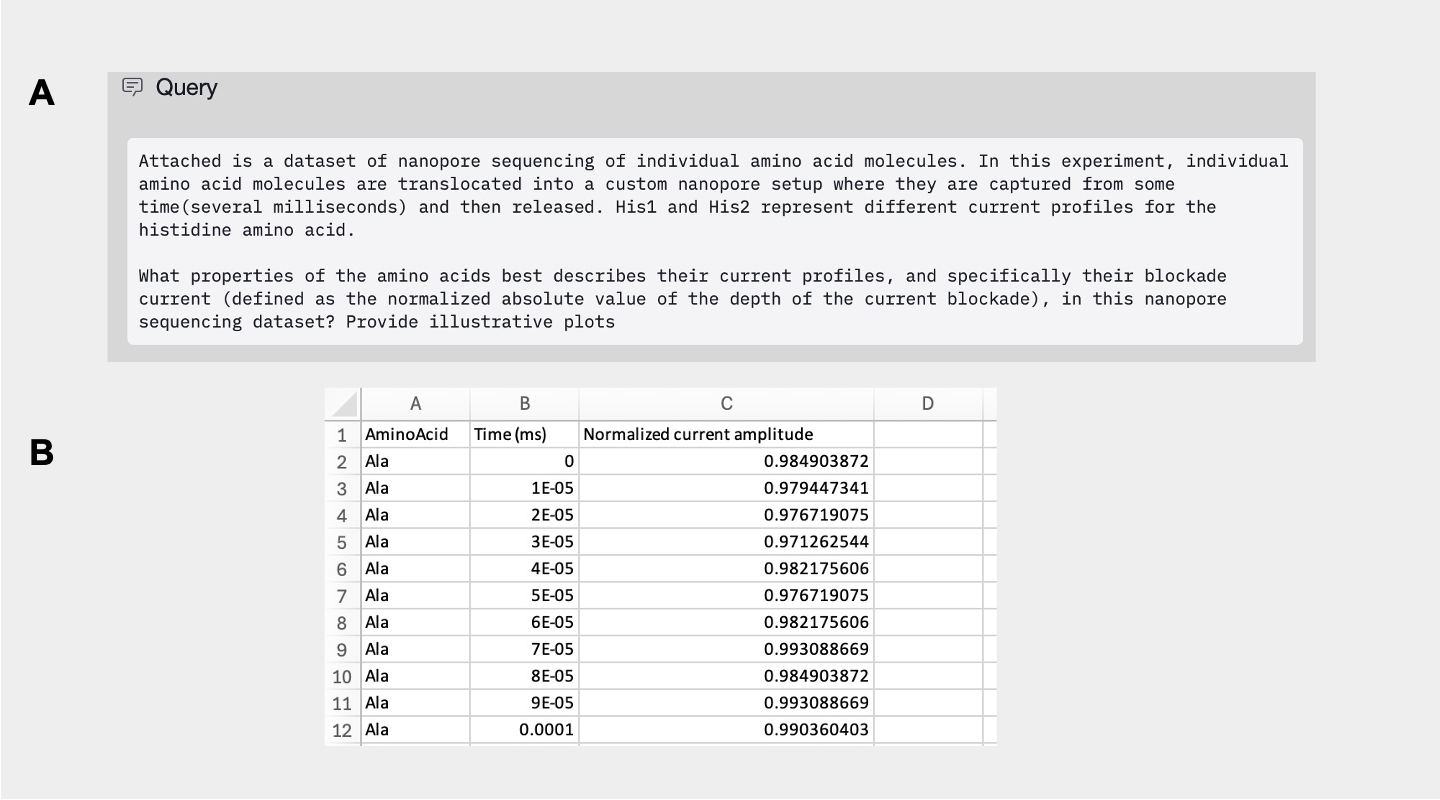
Edison Analysis Agent Determines Current Blockade Amplitudes and Relevant Biophysical Values of Amino Acids
After exploring the dataset, the Analysis agent identified the open pore current (current values when no molecule is present in the nanopore) and the current blockade (current values when a molecule is present in the pore). Using these inferred baselines, the agent computed blockade amplitudes in a manner consistent with the definition provided by Zhang et al. (Figure 2A-C, Analysis Trajectory). The Agent then autonomously retrieved the canonical set of biophysical properties of amino acids (e.g. molecular weight, volume, hydrophobicity, etc.) their values from the literature (Figure 2D).
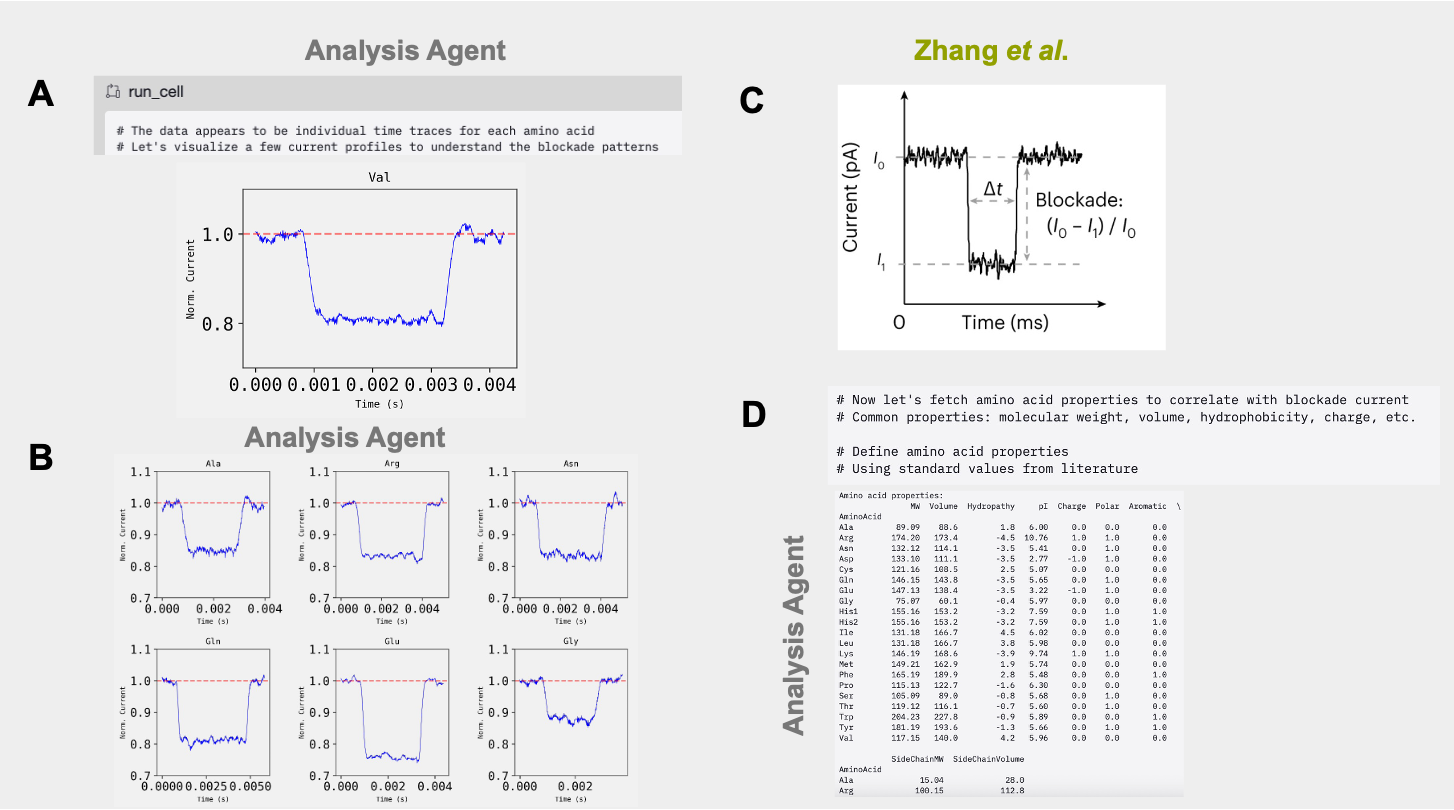
Edison Analysis Agent Identifies Amino Acid Volume as the Main Predictor of Current Blockade Amplitude
The Analysis agent then carried out correlation analysis between the different biophysical properties of amino acids and their blockade current magnitude, identifying volume and molecular weight as the best predictors (Figure 3A, Analysis Trajectory). The blockade current values of amino acids and their molecular volumes are nearly identical to what Zhang et al. showed (Figure 3B), indicating that the Analysis Agent accurately calculated blockade current values and correctly pulled biophysical values of amino acids from the literature. The agent then explored building linear models to predict blockade current using a combination of these biophysical parameters, identifying volume and charge as the most parsimonious set (Figure 3C,D).
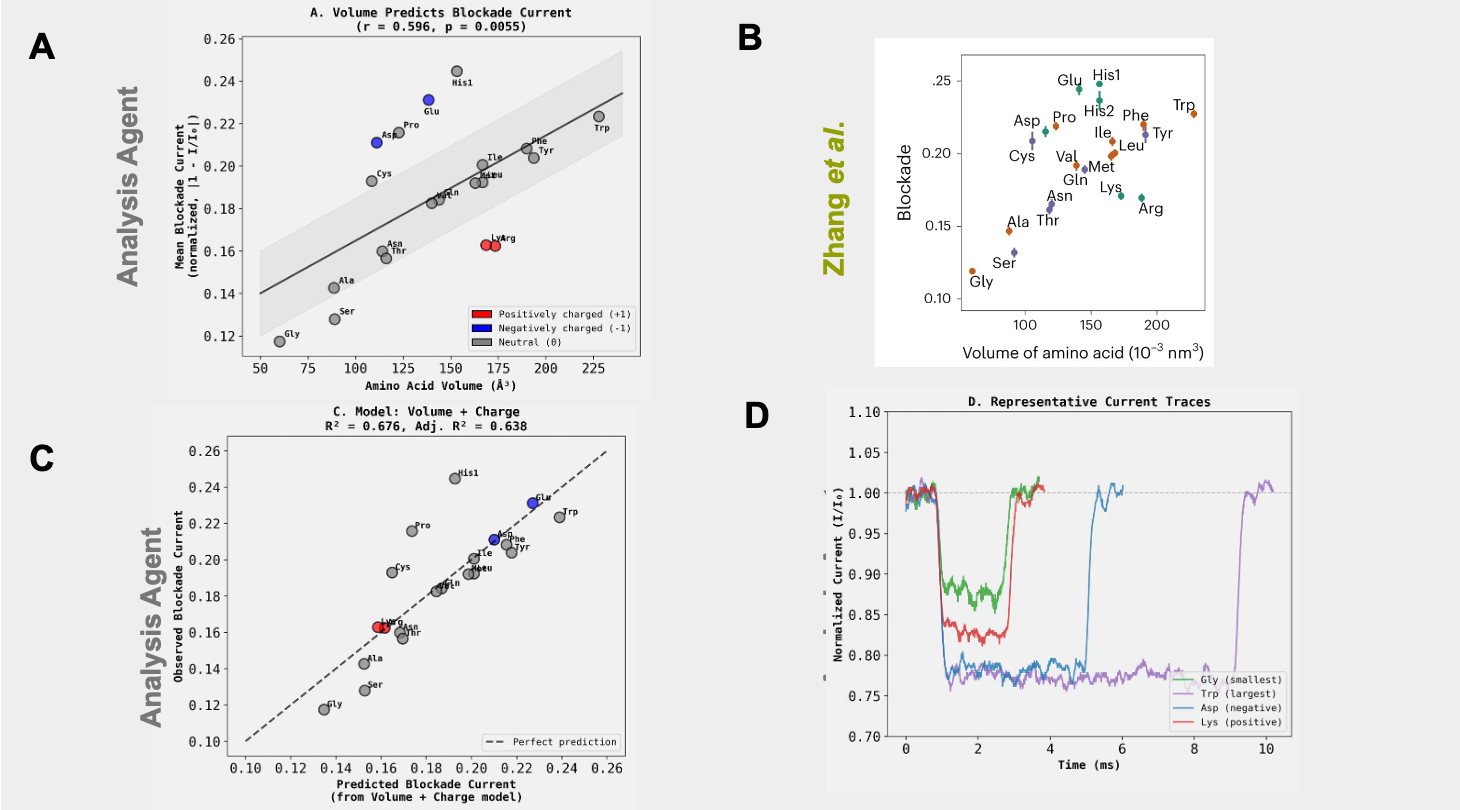
Together, these results demonstrate that the Edison Analysis Agent can autonomously interpret raw biophysical time-series data, extract meaningful quantitative features, integrate external molecular knowledge, and recover a central mechanistic insight from a state-of-the-art nanopore study. Starting from minimally structured current recordings, the agent correctly identified blockade events, computed amino acid–specific current amplitudes, and independently determined that molecular volume is the primary predictor of ionic current disruption within this nanopore configuration, recapitulating a key conclusion of Zhang et al. More broadly, this work highlights the ability of the Edison Analysis agent to move beyond standard bioinformatics workflows into the domain of single-molecule biophysics, where signal interpretation, feature extraction, and mechanistic reasoning must be tightly integrated. As experimental platforms continue to generate increasingly rich and complex biophysical datasets, autonomous systems capable of reasoning across time-series signals and molecular properties will become essential tools for accelerating discovery.




