Edison Analysis Compares Vaccine Design Strategies from Structural Data
Arvis Sulovari
Date:
01.27.2026
Understanding how approved vaccines achieve their efficacy requires integrating structural biology data, protein engineering principles, and clinical trial outcomes—knowledge scattered across PDB structures, UniProt annotations, and published literature. Edison Analysis automates this synthesis with a simple prompt:
"RSV vaccines required stabilizing the F protein in its prefusion conformation to elicit neutralizing antibodies. Retrieve structural and immunological data for prefusion-stabilized RSV F antigens. What design principles enabled this stabilization, and how do the antigen designs in approved RSV vaccines (Arexvy, Abrysvo) differ in their approach and immunogenicity profiles?"
The full trajectory is available here.
Data Retrieval and Integration
In a single run, the agent queried multiple structural and clinical data sources. It retrieved 8 relevant crystal structures from the RCSB Protein Data Bank spanning prefusion and postfusion Respiratory syncytial virus (RSV) F conformations, protein sequence annotations from UniProt, and clinical efficacy data from published Phase 3 trials (AReSVi-006 for Arexvy, RENOIR, and MATISSE for Abrysvo).
The agent identified the foundational DS-Cav1 stabilization technology (McLellan et al., Science 2013) by analyzing PDB structures 4JHW (first prefusion F structure) and 4MMU (DS-Cav1 variant). It extracted the key stabilizing mutations: S155C + S290C (disulfide bond engineering) and S190F + V207L (cavity-filling mutations), and traced the evolution of stabilization approaches through subsequent variants.
Structural Analysis and Vaccine Comparison
The agent synthesized structural principles with clinical outcomes to compare the two approved RSV vaccines. Both vaccines use prefusion-stabilized F protein to display antigenic site Ø (zero)—a critical epitope that elicits potent neutralizing antibodies and is absent in the postfusion conformation.
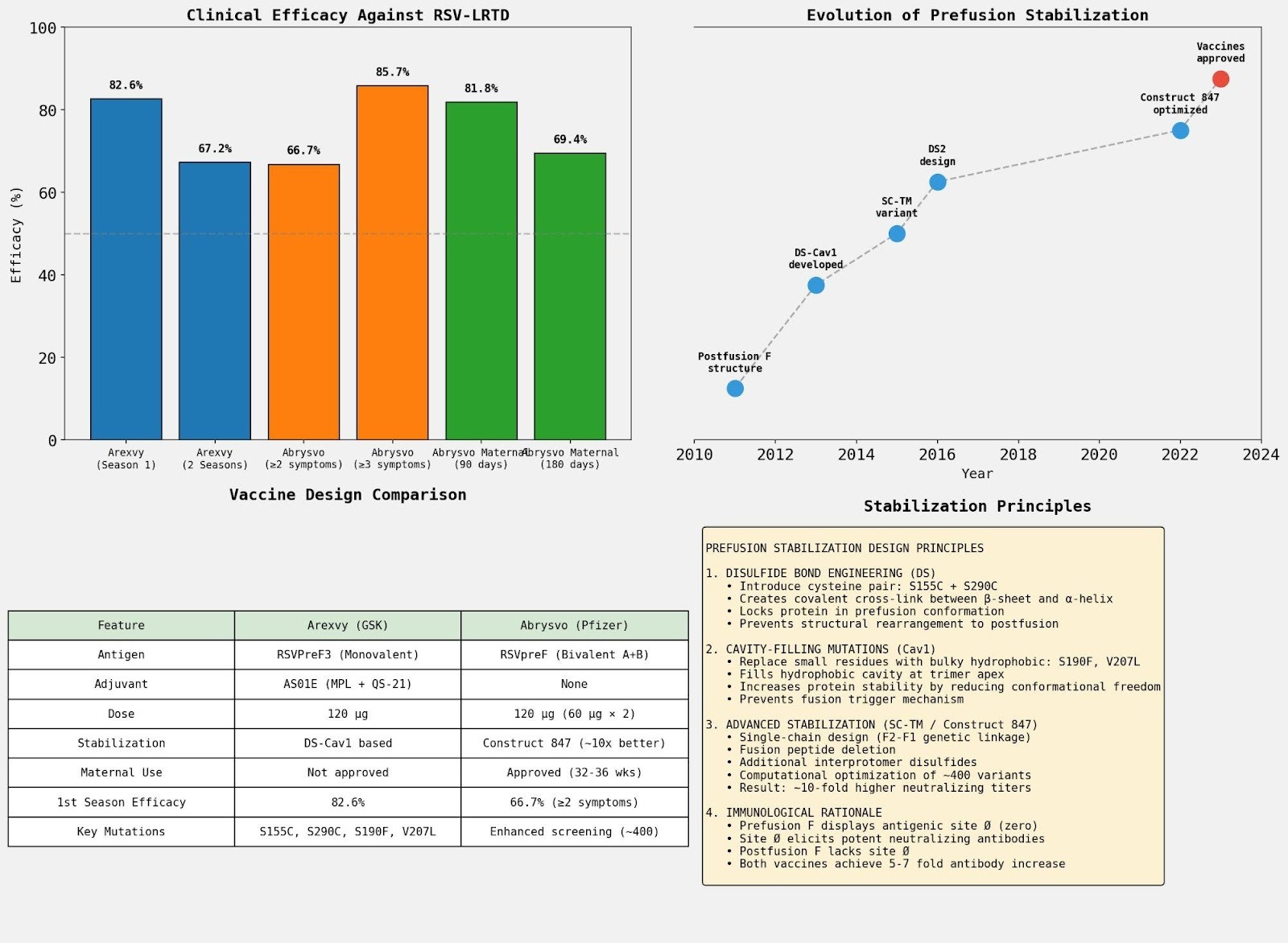
The analysis revealed key differentiators between the vaccines: Arexvy (GSK) uses DS-Cav1-based stabilization with AS01E adjuvant, achieving 82.6% first-season efficacy in adults. Abrysvo (Pfizer) uses the computationally optimized Construct 847 (~10-fold better preclinical neutralizing titers than DS-Cav1) without adjuvant, and is uniquely approved for maternal immunization. Both achieve 5-7 fold increases in neutralizing antibodies sustained for up to 12 months.
Structured, Auditable Outputs
The final output includes a comprehensive summary of stabilization principles, a side-by-side vaccine comparison table, and efficacy data with confidence intervals and trial references. The agent transparently noted limitations including the lack of head-to-head clinical immunogenicity data and differences in trial endpoint definitions that complicate direct efficacy comparisons.

By automating the integration of structural biology data, protein engineering principles, and clinical trial outcomes, Edison Analysis enables researchers to rapidly review how molecular design decisions translate to vaccine efficacy, connecting protein structures to Phase 3 clinical results in a single run.




