Edison Analysis: Data Access, Hypothesis Generation and Testing
Angela Yiu
Date:
02.20.2026
Scientific discovery often starts with an open-ended question. In this example, we demonstrate the capabilities of Edison Analysis to execute a research workflow from data access to hypothesis testing.
We provided the agent with the following high-level prompt:
"Fetch the latest Genomics of Drug Sensitivity in Cancer (GDSC) dataset with IC50s drug screening data across different cancer cell lines. Perform exploratory data analysis to identify any potentially biologically relevant patterns. Based on that, propose an hypothesis and test the hypothesis using an independent publicly available dataset."
The agent then autonomously retrieved and explored the specified data, formulated a biological hypothesis and tested it with an independent dataset. The full trajectory can be accessed here.
Autonomous Data Retrieval & Exploration
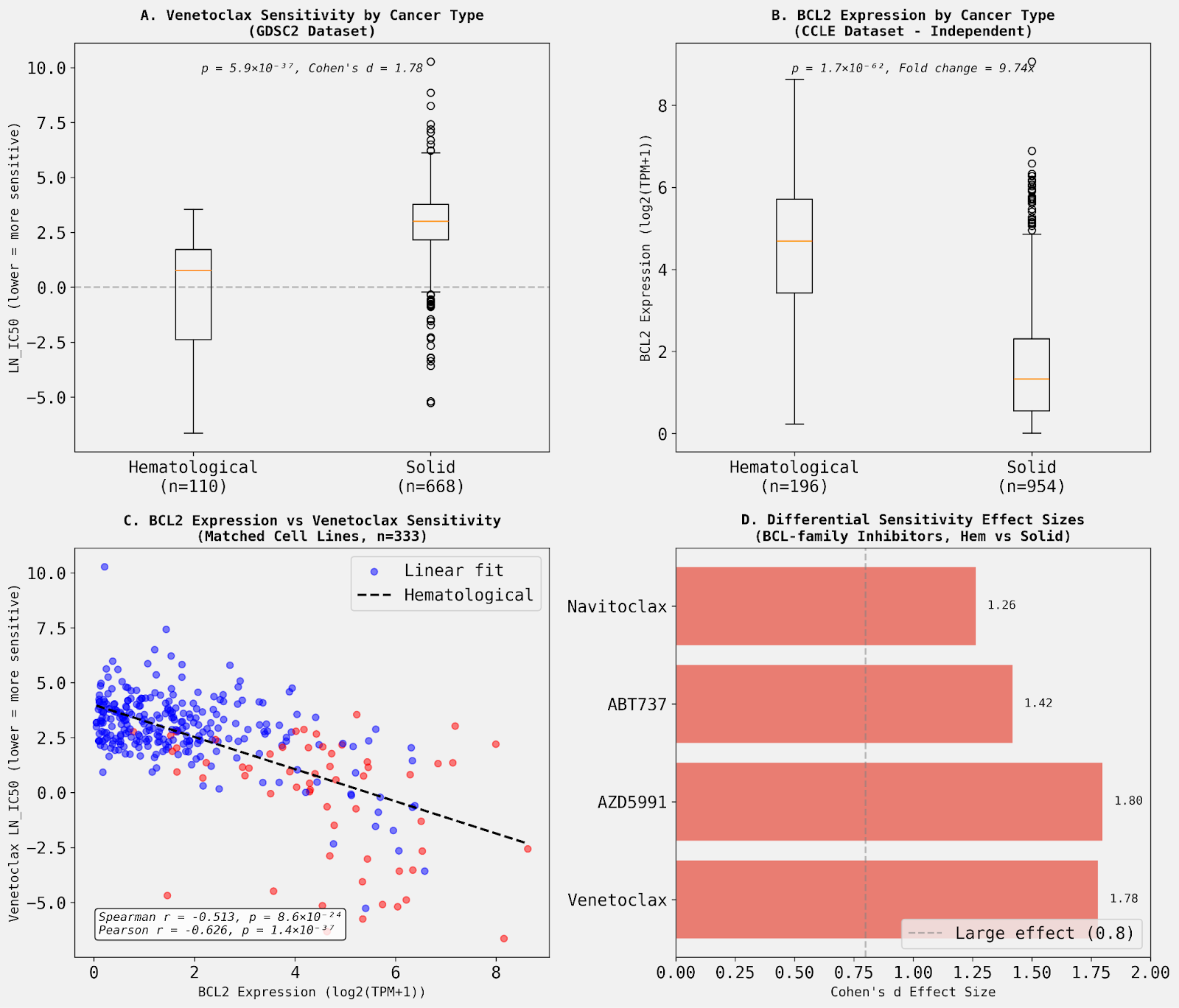
The agent utilized its integrated data access capabilities to retrieve the latest Genomics of Drug Sensitivity in Cancer (GDSC2) dataset, containing 242,036 IC50 measurements across 969 cell lines and 286 drugs (Figure 1). By performing exploratory data analysis (EDA), the agent noticed a significant pattern: hematological cancers (leukemias and lymphomas) exhibited higher sensitivity to BCL-family inhibitors compared to solid tumors (Figure 2A).
Biological Hypothesis Generation
Based on its initial findings, Edison Analysis formulated a biological hypothesis to explain the differential drug response:
- Observation: BCL-family inhibitors, such as Venetoclax, AZD5991, ABT737, and Navitoclax show significantly higher potency in hematological versus solid tumor cell lines (Figure 2A).
- Hypothesis: This sensitivity is driven by higher expression of BCL2 family anti-apoptotic genes in hematological cancers, creating a greater dependency on anti-apoptotic signaling for survival.
Independent Dataset Testing & Validation
To test this hypothesis, the agent retrieved RNA-seq gene expression data from the Cancer Cell Line Encyclopedia (CCLE). By matching 333 cell lines across these two separate repositories, it performed an orthogonal validation:
- Expression Differential: BCL2 expression was found to be 9.74x higher in hematological cancer cell lines than in solid tumors (p = 1.7×10⁻⁶²) (Figure 2B).
- Mechanistic Link: Cross-dataset correlation confirmed that higher BCL2 expression is independently associated with increased Venetoclax sensitivity (r = -0.626, p = 1.4×10⁻³⁷), even when controlling for cancer type (Figure 2C).
- Biological Interpretation: Hematological cancers express ~10-fold higher BCL2 levels than solid tumors, creating "oncogene addiction" to anti-apoptotic signaling. BCL-family inhibitors such as Venetoclax exploit this dependency. This aligns with the fact that Venetoclax is FDA-approved for hematological malignancies (CLL, AML) but not for solid tumors.
Conclusion
This workflow demonstrates the agent’s ability to autonomously fetch specified datasets, explore them for non-trivial patterns, and test self-generated hypotheses against independent data. By automating the heavy lifting of data engineering and statistical validation, Edison Analysis allows researchers to arrive at validated pharmacogenomic biomarker relationships in a fast and traceable manner.
While this demonstration highlights a focused investigation on one aspect of the dataset, our platform is capable of even greater complexity. For large-scale, in-depth investigations, we recommend using Kosmos to expand the depth and breadth of discoveries. Here is an example of a Kosmos run with the same prompt where it also identified the same trend, but explored much deeper.




