Expanding Kosmos' Data Access Capabilities
Angela Yiu, Ludovico Mitchener
Date:
01.14.2026
Kosmos now integrates with the most common scientific external data sources, from Zenodo to the Protein Data Bank, covering 80% of all scientific data commonly used by the life science community.

Previously, researchers had to manually curate, identify and download suitable datasets prior to a Kosmos run. Kosmos can now autonomously search and retrieve the most suitable publicly available datasets based on your research objectives. Users do not need to prepare and upload public data to initiate a Kosmos run. This is available today on our platform.
Kosmos can fetch relevant public data at various stages of the research process:
- Project initiation: identifying suitable public datasets to address your research objectives
- Data enrichment: supplementing your datasets with relevant public data during the research process to address any gaps, improve statistical power and confidence in the results
- Finding validation: validating Kosmos’ findings by cross-referencing against independent, orthogonal datasets
Investigating TGFβ Signaling in Pancreatic Cancer
To demonstrate these capabilities, we provided Kosmos with a single in vitro dataset (RNA-seq from TGFβ-treated PANC-1 cells) and a broad research objective to investigate pancreatic ductal adenocarcinoma (PDAC) tumorigenesis across different modalities and models. Kosmos autonomously navigated the rest.
User input:
Research Objective: Investigate how TGFβ-induced molecular signatures contribute to PDAC tumourigenesis in in vitro cell and in vivo mouse models. Validate your main findings with relevant clinical data.
Data description: These are RPKM data from RNAseq of PANC-1 cells that were treated with 5nM TGFβ or vehicle control for 72h.
Here's how Kosmos leveraged public data at each stage:
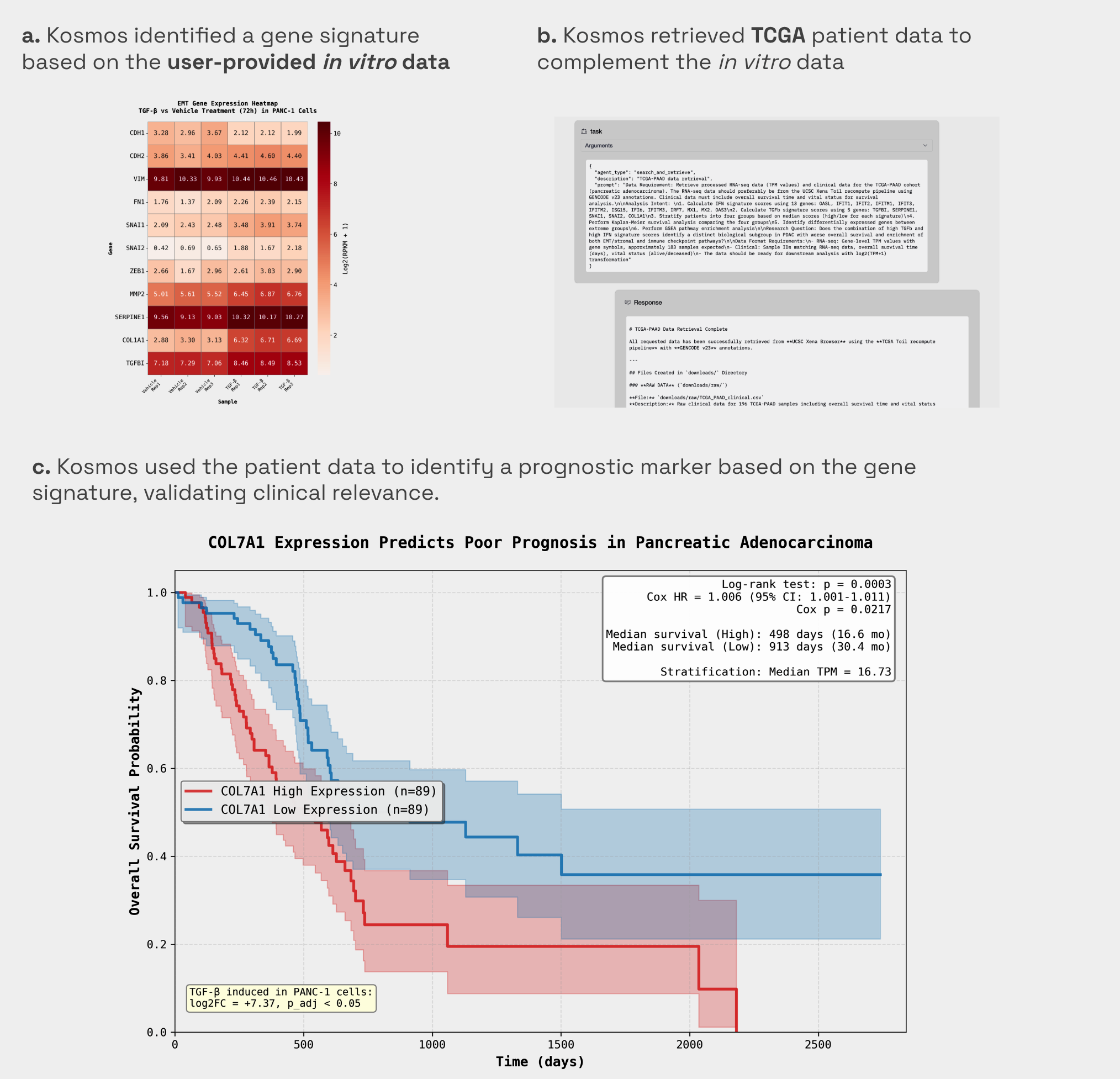
1. Project Initiation: Connecting In Vitro Gene Signature to Patient Survival
After identifying the extracellular matrix (ECM) as a top TGFβ-inducible gene signature from the user's data [1. Figure a], Kosmos autonomously searched for and retrieved clinical bulk RNA-seq and survival data from the TCGA-PAAD cohort [2, Figure b]. It successfully identified COL7A1, part of the ECM signature, as a potent prognostic marker, finding that high expression predicts significantly worse outcomes, with a median survival of 498 days compared to 913 days for low expression (p=0.0003) [3, Figure c]. This step allowed the run to move from a petri dish observation to a clinically relevant human disease finding without any manual data entry.
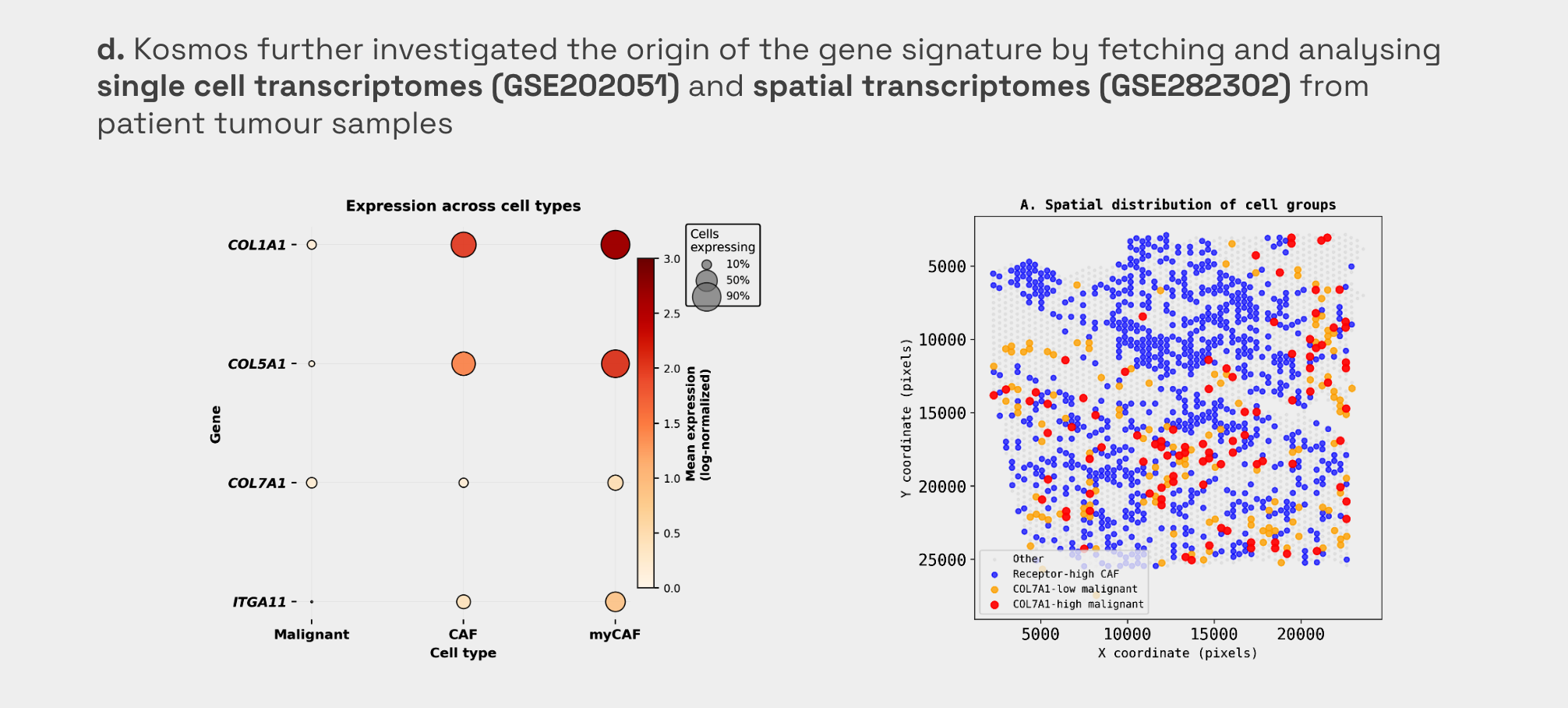
2. Data Enrichment: Resolving Cellular Origins with Single-Cell RNA-seq
To investigate the mechanisms behind this signature that correlates with poor survival, Kosmos enriched the project by fetching single-cell transcriptomes (GSE202051) from patient tumor samples [4]. The analysis revealed a "division of labor" that bulk data could not show: while most ECM genes (COL1A1, COL5A1) were produced by fibroblasts, COL7A1 was primarily sourced from the malignant epithelial cells themselves (66.2%) [5, Figure d]. This data enrichment allowed Kosmos to propose a specific mechanism: a paracrine feed-forward loop where tumor cells and fibroblasts "talk" to each other to drive invasion.
3. Finding Validation: Proving Paracrine Signaling with Spatial Context
Finally, Kosmos validated the proposed paracrine interaction by retrieving an orthogonal spatial transcriptomics dataset (GSE282302). It performed a proximity analysis which confirmed that COL7A1-high malignant cells are physically located significantly closer to receptor-positive fibroblasts than COL7A1-low cells (p = 0.0052) [6. Figure d]. This spatial evidence served as an independent validation of the mechanistic findings derived from the previous single-cell and bulk datasets.
What this unlocks
With autonomous public dataset retrieval, a Kosmos run can now begin with just a single dataset or even just a research objective. It can autonomously expand into a multi-modal evidence chain— clinical data validation, mechanistic investigation via single-cell and spatial context—without any manual dataset hunting or downloading.




