Introducing Edison Analysis
Ludovico Mitchener, Jon Laurent, Angela Yiu, Arvis Sulovari, Conor Igoe, Alex Andonian
Date:
11.20.2025
Today, we are announcing Edison Analysis, our next-generation scientific analysis agent.
Today, we are announcing Edison Analysis, our next-generation scientific analysis agent. Edison Analysis is a major evolution of FutureHouse’s Finch, which was previously only available via closed beta.Edison Analysis is also the analytical engine underpinning Kosmos, our AI Scientist. It is available today on our Platform and is also usable via our API.
Edison Analysis performs complex scientific data analysis tasks by iterative updating of Jupyter notebooks in a dedicated environment. Given datasets and a prompt, the agent systematically explores, analyzes, and interprets the data to provide comprehensive answers and insights. Key features include:
- SOTA performance across key scientific data analysis and access benchmarks.
- Science-native design: It works with Python, R, and Bash code execution and iteratively builds Jupyter notebooks to answer research questions, just like a human scientist.
- Dedicated data access: A specialized tool allows it to access external datasets to supplement its analysis.
- Domain generalization: While it specializes in bioinformatics analysis, it generalizes to all analytical domains.
- Environment: It is built with a Docker image including the most common scientific data analysis packages.
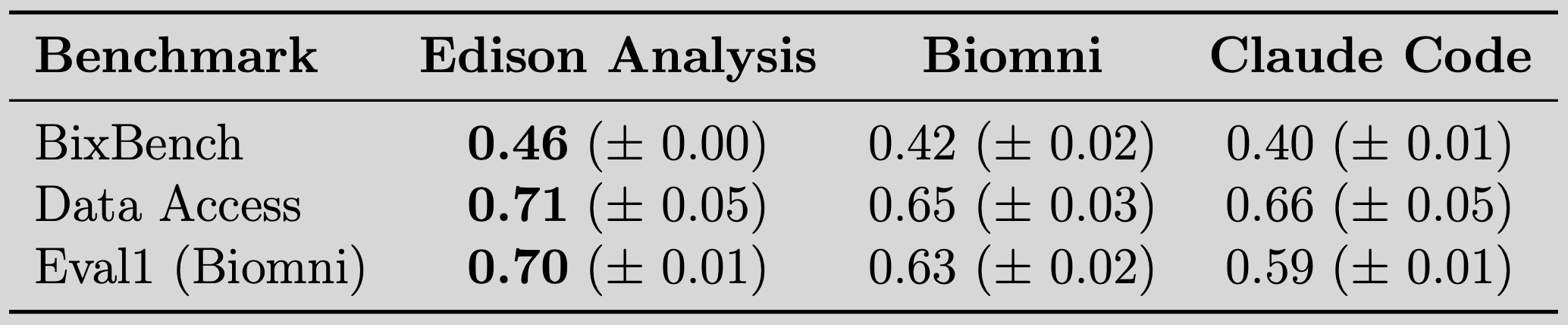
We've measured performance for Edison Analysis across key benchmarks for analyzing and accessing data, and found that it outperforms other available systems:
Science-native
Edison Analysis is a science-native coding engine. While modern software engineers have a plethora of AI tools to support their work, we wanted to build a coding tool that prioritizes the iterative, exploratory nature of scientific analysis.
To achieve this, we moved away from opaque file systems and hidden logic. Edison Analysis operates entirely within a Jupyter notebook, where it iteratively edits cells. This design choice ensures that every step of the scientific process is transparent. The reasoning trace is fully auditable within the notebook, allowing users to see exactly how data was manipulated, which parameters were chosen, and how conclusions were drawn. The agent’s results are shared in a clear Markdown report, and it always generates a figure to visually summarize its findings.
This design also ensures the analysis is not locked to our platform. Users can ask follow-up questions to the agent, but they can also download the notebook and its associated data to their local environment at any point. This facilitates easy hand-offs between the agent and the scientist; for example, a researcher can download the agent's output to manually refine plotting configurations for a final manuscript, or to extend the analysis locally without needing to re-engage the agent. We chose the Jupyter notebook for its simplicity, approachability, and ubiquity in the scientific community.
We measured Edison Analysis performance on analytical tasks with a purpose-built benchmark called BixBench, released by FutureHouse earlier this year, which has become a standard evaluation for any bioinformatics agent system. Edison Analysis outperforms two available comparator agents, Biomni and Claude Code, on BixBench, reaching 46% overall accuracy (see table above.)
We also measured performance of a panel of human bioinformatics experts on a subset of BixBench capsules pre-selected to cover the spectrum of difficulties. We compared this human performance to Edison Analysis, and found that the agent nearly matched human experts in average performance across the selected capsules, while actually besting humans on two individual capsules.

Accessing exernal data
In scenarios where the agent needs data to supplement its analysis, it utilizes search_and_retrieve, a dedicated tool for obtaining data from various scientific APIs. Users often use Edison Analysis to search and select a dataset to feed to Kosmos, our AI scientist.
This tool provides access to the most common scientific external data sources, from Zenodo to the Protein Data Bank. It features specialist connectors for the top 20 most frequently used sources, and can also perform one-shot API queries for many other databases by leveraging web search to build documentation and structure queries.
Prior to the integration of this tool, we found that a significant amount of time (and a large portion of the context window) was wasted attempting to retrieve data from complex or convoluted APIs. The search_and_retrieve tool streamlines the agent’s access to data, ensuring the context window is reserved for performing deeper analysis.
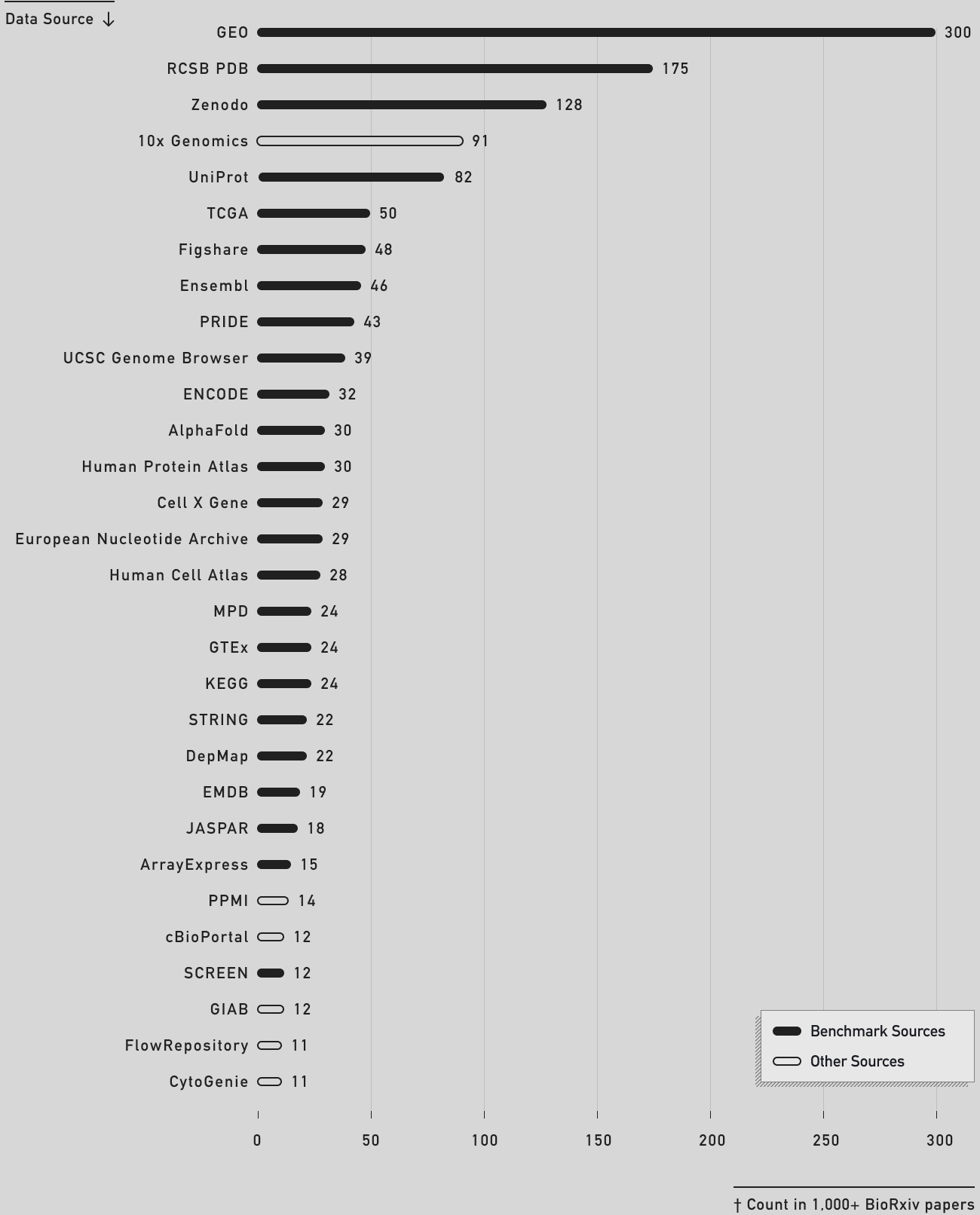
We measured Edison Analysis' access to important data sources using a dedicated Data Access Benchmark that will be released in the coming weeks, and compared its performance to Biomni and Claude Code (see table above). To build the benchmark, we analyzed a set of over 1,000 random papers posted this year in bioRxiv for the data sources they utilized, and then had subject matter experts manually filter them for priority, credentialed access issues, redundancy, or other issues. In the chart below, you can see which data sources from the top 30 sources in bioRxiv papers are included in our benchmark.
We also evaluated Edison Analysis on an external benchmark from Biomni, Eval1, a significant portion of which we characterize as a data access or retrieval benchmark. Eval1 includes a large proportion of tasks from FutureHouse’s LAB-Bench, including a set from the database access subset DbQA. Edison Analysis surpasses both Biomni and Claude Code on this benchmark, reaching 70% accuracy.
Demos
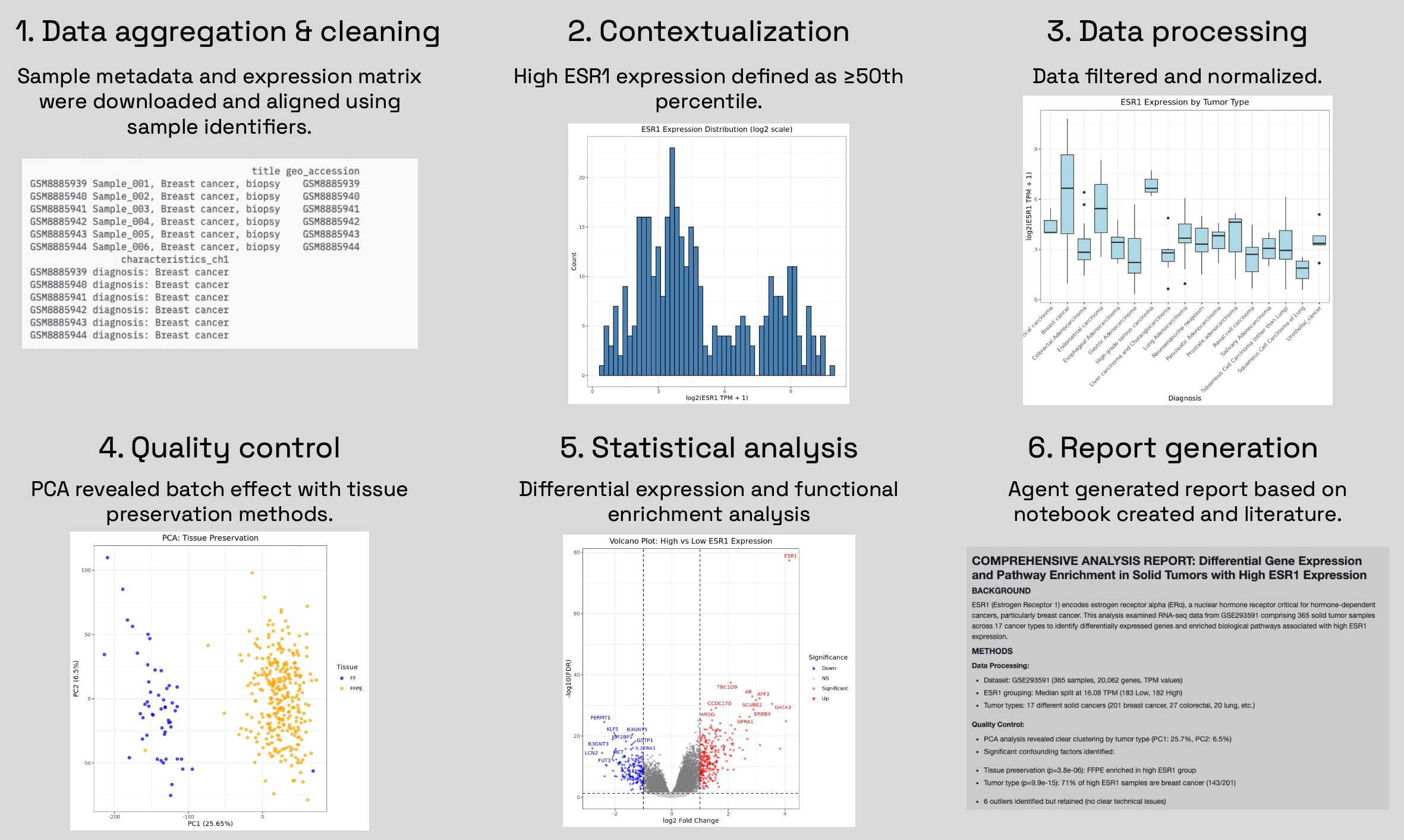
In a single run, Edison Analysis can perform data analysis workflows end-to-end, from data aggregation to report generation that would typically require a skilled bioinformatician several hours. As an example, we asked Edison Analysis to fetch RNAseq data from GEO accession GSE293591, identify differentially expressed genes and enriched biological pathways in solid tumors with high ESR1 expression, and extract biologically relevant insights from the results. The agent identified nuances in the data, such as skewed distribution of ESR1 expression, cancer type imbalance, and batch effect due to tissue preservation methods, and addressed them accordingly.
Edison Analysis can also be applied across a wide range of use cases (click on each to view the full agent trajectory on the Edison Platform):
- Parameter sweep: Determining the most appropriate resolution for scRNAseq clustering
- Data harmonization: Analysing post-transcriptional control in Parkinson’s Disease in R
- Replication analysis: Assessing eQTL from GTEx in GEUVADIS in R
- Survival analysis: Identifying microRNA signature that can predict survival
- Metabolic modeling: Evaluating maximum mixotrophic growth with flux balance analysis
- Lab data analysis: Assessing specificity and efficiency of qPCR primers
Thanks
Many people contributed to building Edison Analysis. Ludovico Mitchener led the design and engineering with significant support from Edwin Melville-Green, Alexander Andonian, Siddharth Narayanan, James Braza, Michael Skarlinski, and Christopher Zou. Angela Yiu led academic collaborations and provided critical feedback on Edison Analysis with significant support from Arvis Sulovari. Alexander Andonian led Edison Analysis performance optimizations with significant support from Conor Igoe. Jon Laurent led and supervised all benchmark creation and human evaluation efforts, with significant support from Alex Andonian, Conor Igoe, Sam Cox, and Zachary Siegel. Andrew White and Sam Rodriques together supervise technical work at Edison Scientific.




