LABBench2: An Improved Benchmark for Measuring AI in Biology Research
Jon Laurent
Date:
02.05.2026
Edison Scientific’s primary objective is to automate scientific discovery, and we take seriously the need to reliably measure progress towards that goal. Previously, we released benchmarks to measure this progress: LAB-Bench was the first agentic benchmark for research task capabilities, moving beyond pure knowledge to real-world work. BixBench was the first dedicated benchmark for measuring the bioinformatics skills of AI systems. Our ether0 model was the first chemical reasoning model, and came along with a dedicated benchmark for measuring its abilities.
We now release an evolution of LAB-Bench, which we (unsurprisingly) call LABBench2. LABBench2 covers similar broad categories to LAB-Bench, while adding a couple of important new ones. In sum, we are releasing 1,892 tasks across 11 broad categories.
Rethinking Practical Evaluation
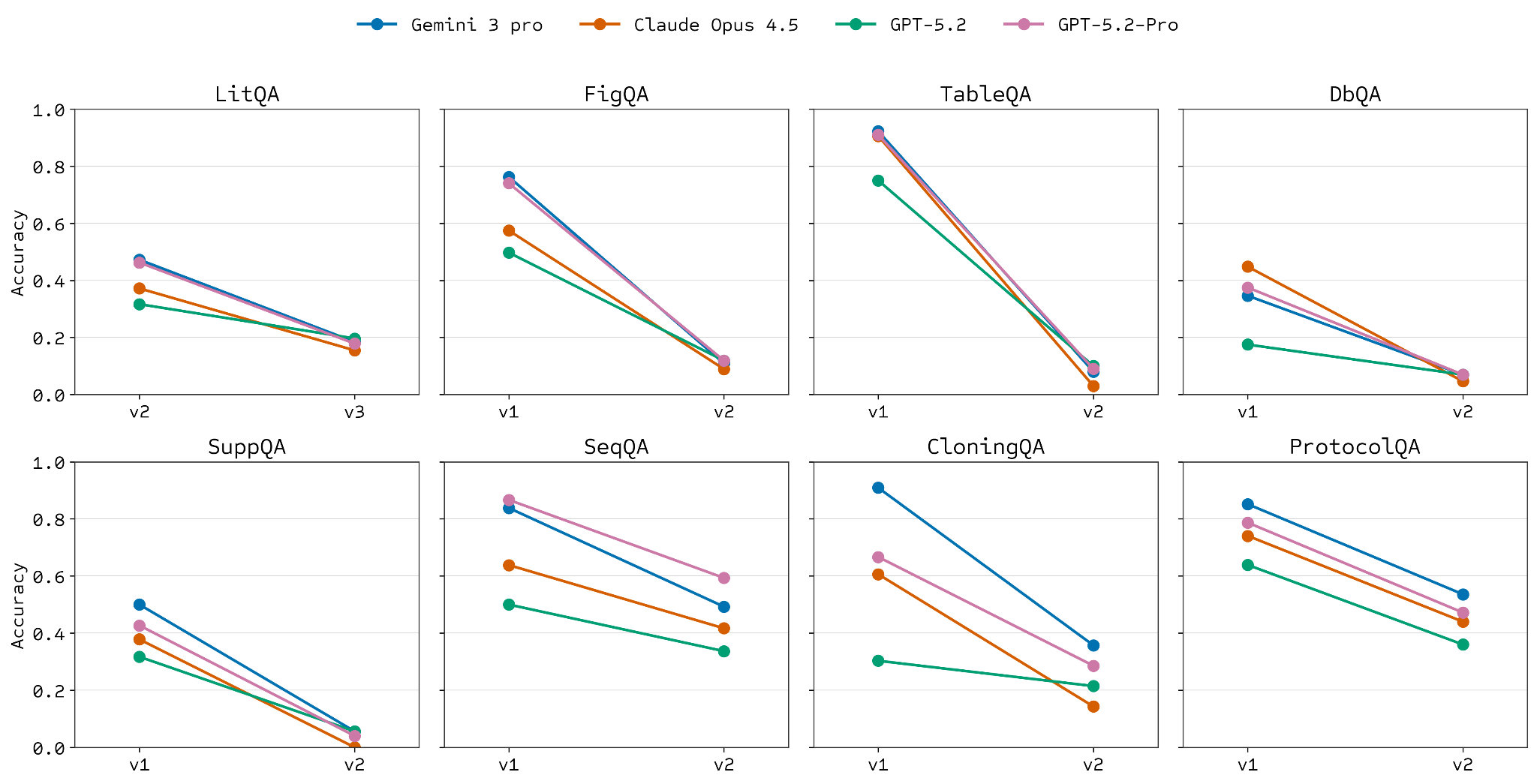
We revise and rethink our approach to task design across all categories, with a particular emphasis on reliable retrieval as a key need for successful discovery research. Where LAB-Bench included tasks, like FigQA or TableQA, that required multi-modal capabilities, LABBench2 combines this with the need to retrieve and “read” the correct source text and figures. We similarly add modifications for SeqQA2 and CloningQA that require retrieving relevant sequences to answer. Importantly, in both cases, we can also provide the necessary context in-line or in a local file, to delineate retrieval-limited performance.
We also increased the specificity and scope of database access tasks for DbQA2, so we are benchmarking against more (and more important) data sources. We do the same for SuppQA2, where we include requirements to access not just supplemental text but also figures, tables and alternative files.
In addition to revamping the approach to the original LAB-Bench task types, we add a few important new tasks. High-value applied research relies on other sources of information, including patents and clinical trials. We thus add PatentQA and TrialQA to the suite to address these needs. Importantly, these new tasks are in part multi-modal, sometimes requiring reading a figure or table in a patent or trial. Discerning the most relevant or reliable sources for a given line of inquiry is becoming increasingly important as agents are more capable of deep research. To address this, we’ve introduced another new task category, SourceQualQA. Here, we evaluate whether models can discern why a certain research study was not included in a systematic review of a specific topic.
What are the results?
One of the goals we set when deciding to build LABBench2 was to make it more difficult than the original. Despite some LAB-Bench subsets still being difficult, we have seen saturation or superhuman performance on others [1, 2]. As shown in the plots below, we have universally achieved this goal across like-for-like categories using current models.
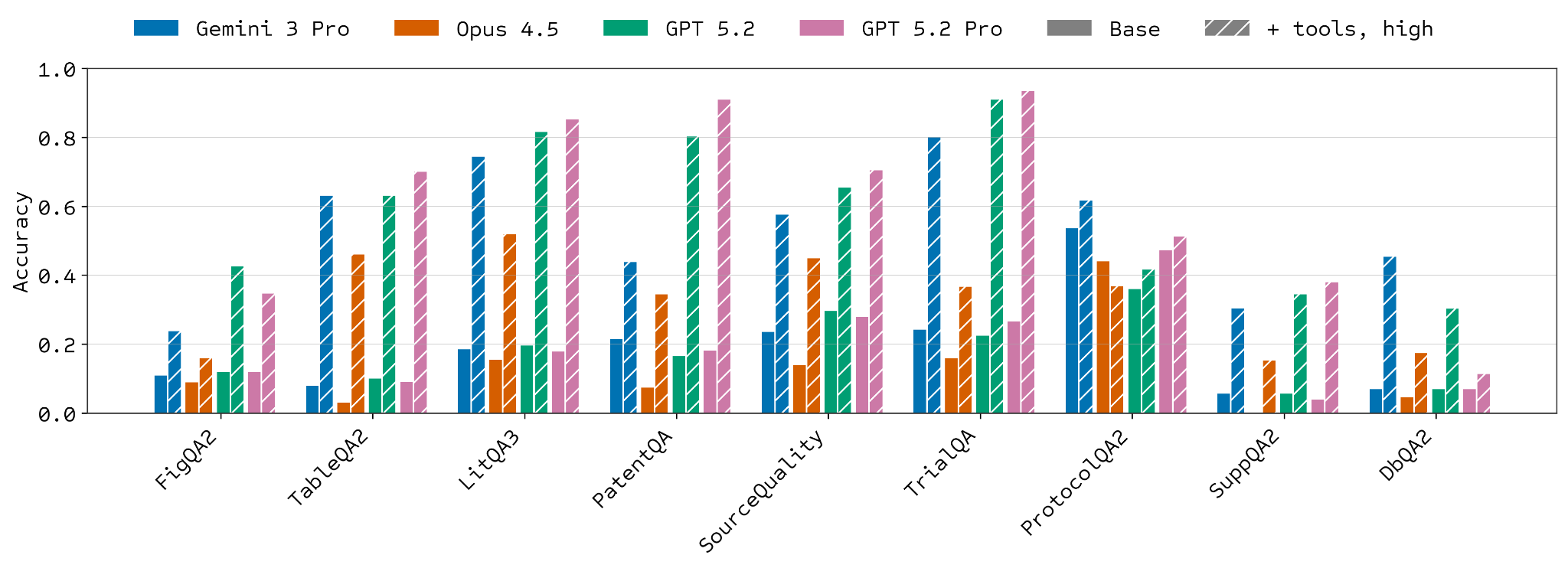
Despite this increased difficulty, one of the key improvements driving model performance over the last couple of years has been the addition of sophisticated tools, such as web search and code execution, to base models. Both are particularly relevant for LABBench2 tasks. A dramatic (though variable) increase in performance is observed when models are allowed to use these tools. This most obviously applies to web search on retrieval tasks but is also beneficial for sequence manipulation tasks like SeqQA2 and CloningQA, presumably due to code execution capabilities.
Limitations and the Future
With LABBench2, we’ve addressed many of the limitations built into LAB-Bench, such as eliminating multiple-choice questions, increasing the realism across all tasks, and providing mechanisms to delineate specific limitations, like retrieval or file handling. LABBench2 still necessarily includes its own concessions, especially in the scope of both individual tasks and of the categories of tasks covered.
We are steadfast in our commitment to defining the frontier of scientific benchmarks and are particularly excited about the opportunities ahead, both i) to build deeper, more thorough and pointed benchmarks in specific areas of focus (BixBench being an initial but still somewhat lacking foray into this for bioinformatics) and ii) to build higher-level evaluations for more complex autonomous systems. For example, building frameworks for evaluating systems like Kosmos that can carry out long-horizon, autonomous discovery campaigns.
Each of these areas should also be built with increasingly real-world context, for example, by moving evaluation into the physical wet lab and focusing on high-value applied research areas like drug development.
If you are interested in joining us to work on evals like these, please get in touch at evals@edisonscientific.com or see our open roles here. Likewise, if you have ideas for evals we should build or would like to work with us to leverage our pipeline to build specific evals, please get in touch at evals@edisonscientific.com.
Get the paper PDF.
See the dataset.
Use the harness.




