Single Cell Analysis Reveals the Expansion of T-cells in mRNA Neoantigen Vaccine Treatment of Pancreatic Adenocarcinoma Patients
Asmamaw (Oz) Wassie, Michaela Hinks
Date:
01.30.2026
Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal cancer that has long been considered refractory to immunotherapy because of its low mutation burden and immune-excluded tumor microenvironment. In a landmark study, Rojas et al. challenged this view by showing that a personalized mRNA neoantigen vaccine (autogene cevumeran), generated from resected PDAC tumors and administered with anti-PD-L1 therapy and chemotherapy, can elicit strong neoantigen-specific CD8⁺ T-cell responses in a substantial fraction of patients. These vaccine-induced T cells were durable and their presence strongly correlated with delayed disease recurrence, providing compelling evidence that individualized neoantigen vaccination can drive clinically meaningful immunity in PDAC. Here we demonstrate that the Edison Analysis agent can autonomously process data and recapitulate a key finding when provided with a single cell RNA sequencing (scRNA-seq) dataset generated by this study. This publicly available dataset consists of paired scRNA-seq and T-cell receptor (TCR) sequencing datasets of T-cells from Peripheral Blood Mononuclear Cells (PMBCs) derived from 5 patients with PDAC treated with a personalized mRNA neoantigen vaccine. Specifically, the dataset consists of preprocessed hdf5 files of cell x gene count matrix along with associated meta data and paired TCR sequencing files of reconstructed TCR sequences and associated cell barcode information.
Edison Analysis Agent Recapitulates Results Showing Vaccine Induced Expansion and Activation of Cytotoxic T-Cells
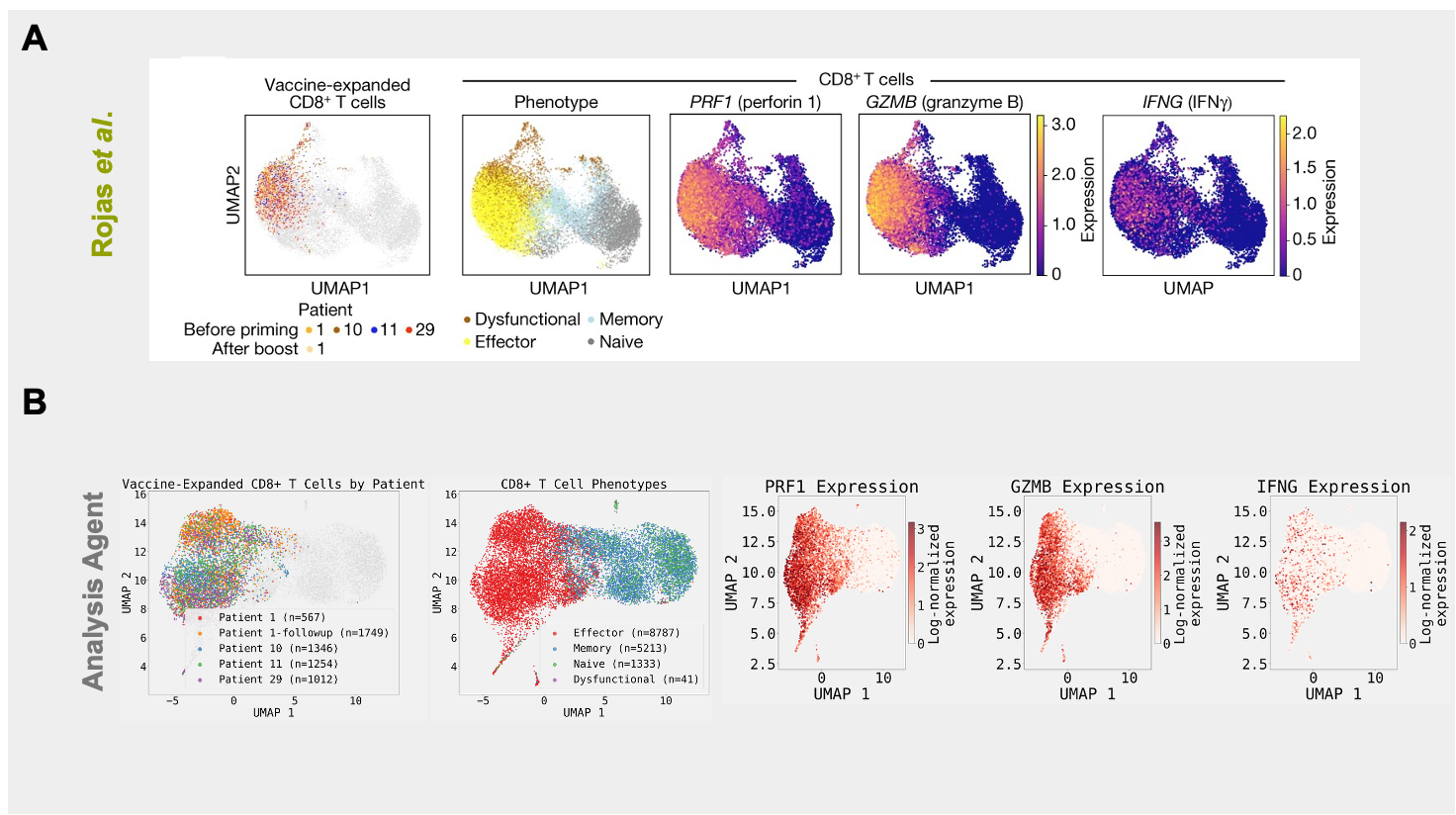
We primarily sought to reproduce a finding from the paper (Figure 2F of manuscript) that the administration of personalized mRNA neoantigen vaccines results in the expansion and activation of Cytotoxic (CD8+) T-cells. We first provided the Edison Analysis agent directly with the paired scRNA-seq and TCR-sequencing files for each patient, along with a prompt instructing the agent with the analysis steps to carry out to reproduce the finding in the paper (Figure 1). The Analysis agent first aggregated the datasets as instructed and applied quality control (QC) steps to set a minimum number of genes expressed per cell cutoff and to filter out mitochondrial reads. After, the agent used Harmony for batch correction using samples as batch labels on 50 principal components (from Principal Component Analysis it spontaneously carried out) and 1521 highly variable genes it identified. Having then identified CD8+ T-cells based on CD8A and CD8B expression levels, the agent searched the literature to curate marker genes for CD8+ T-cell phenotype and applied them to classify these cells based on their state (i.e. effector, naive, memory, or dysfunctional). Finally, the agent used TCR sequences to quantify clonal expansion.

The Edison Analysis agent was able to recapitulate the main findings the authors derived from this dataset. In their work, the authors demonstrate that vaccine-expanded T-cell clones adopt a cytotoxic effector CD8⁺ T-cell state (Figure 2A). Even more, these cytotoxic T-cells express lytic agents, like perforin 1 and granzyme B, meaning that these cells are activated to attack their targets. Similar to the paper, the analysis carried out by the agent shows that the vast majority of the patient-derived vaccine-expanded T-cells have an effector phenotype (Figure 2B, Analysis Trajectory). In addition, these T-cells are also the ones that express the same lytic agents indicated in the paper. The accuracy of the analysis steps of the Analysis agent is apparent in the similarity of the UMAP renderings to the ones shown in the paper. For example, in both cases, effector and naive T-cells are the most transcriptionally distinct phenotypes, while memory T-cells are still distinct but overlap slightly with naive T-cells. Some areas of difference: while the authors show dysfunctional T-cells being more similar to effector T-cells, the UMAP visualization by the Analysis agent shows them being more transcriptionally similar to naive and memory T-cells. In addition, a slightly higher fraction of CD8+ T-cells are identified as expanded by the Analysis agent. These differences likely arise from differences in analysis steps between the authors and the Edison Analysis agent.
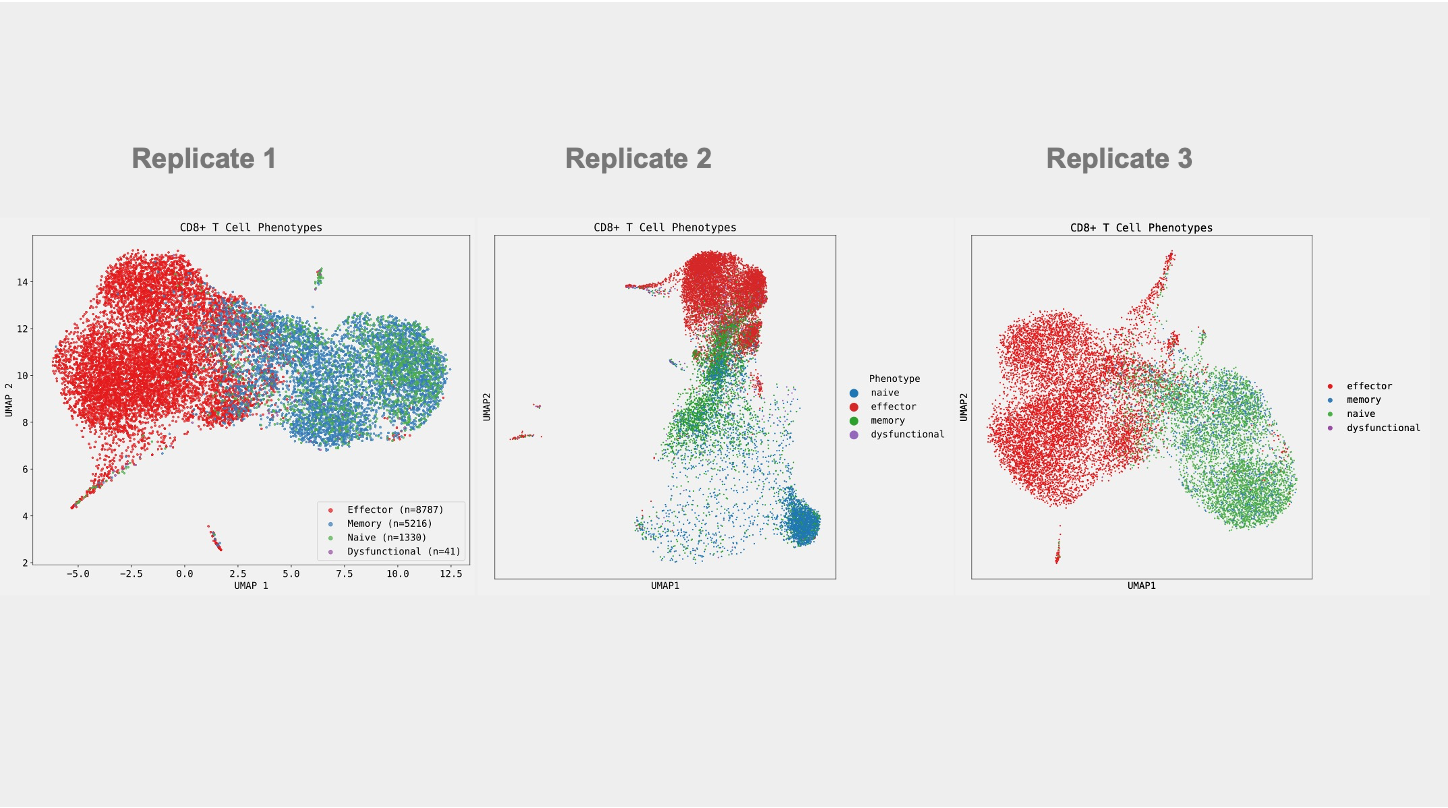
Replicate Runs of Edison Analysis Agent Yield Consistent Results
A key requirement of an autonomous AI Scientific Agent is the ability to yield consistent results when provided the same inputs. To assess consistency, we ran the agent three times providing it the same prompt and dataset as above. Each time, the Analysis agent produced results that retained the same essential biological conclusions (Figure 3, Trajectory 1, Trajectory 2, Trajectory 3). For instance, in each case over 90% of vaccine expanded CD8+ T-cells are classified as having effector phenotypes (91.6%, 96%, 97.6%). In all cases, effector and naive T-cells are the most dissimilar in terms of expression profiles while memory T-cells lie somewhere in between. In addition, the agent used the same criteria for classifying CD8+ T-cells by using the combined expression of CD8A and CD8B and comparing it to the CD4 expression. Yet, there are differences in processing that introduce variability in the final result. Given that the Analysis agent was provided discretion to come up with criteria for classifying CD8+ T-cells, it used slightly differing combinations of marker genes for assigning phenotypes in each run. In addition, the number of highly-variable genes used for the PCA, batch-correction, and UMAP rendering steps are also slightly different. If desired, further consistency can be achieved by providing more narrow instructions to the agent.
In conclusion, when provided instructions to carry out a single cell transcriptomic analysis of an scRNA-seq dataset, the Edison Analysis agent recapitulated a key finding from the Rojas et al. groundbreaking work, that the administration of personalized mRNA neoantigen vaccines results in the expansion and activation of Cytotoxic (CD8+) T-cells. Furthermore, the agent consistently yielded similar results when prompted multiple times with the same dataset and instructions. Though the Analysis agent did not access the manuscript in its trajectories, the text may be present in the training data used by the LLM agent. While some of the analysis steps were concretely defined in the prompt, the agent made reasonable bioinformatic decisions where it was provided discretion. Overall, this exercise demonstrates the ability of the Edison Analysis agent to accurately derive biological insights from single cell transcriptomic datasets.




